|
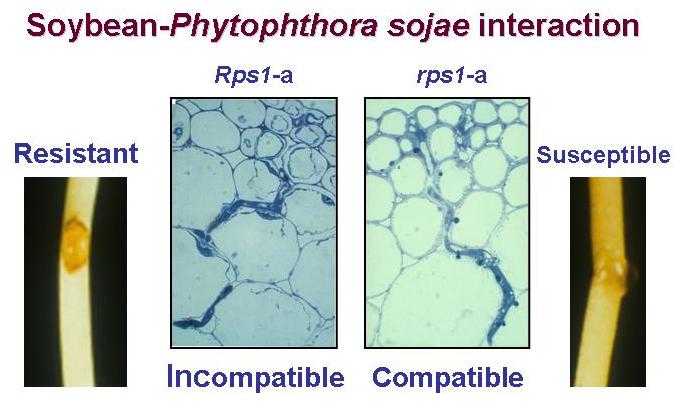
Understanding the molecular basis of the soybean-P. sojae interaction is the primary goal of the Bhattacharyya lab. The lab investigates two important aspects of this interaction: i) the recognition and ii) the signal transduction. These processes activate defense genes involved in the synthesis of anti-microbial defense compounds following infection. The recognition process, determined by the products of a pair of corresponding genes from soybean and P. sojae, presumably activates a signal pathway leading to the expression of defense genes. |
 |
|
The lab has applied multiple molecular approaches in understanding molecular basis of the soybean- P. sojae interaction. (i) A map based cloning approach was applied in cloning the soybean disease resistance genes Rps1-k that determines the recognition process. (ii) They have applied a yeast two-hybrid approach in isolating Rps1-k-intarcting proteins to better understand the signaling factors involved in the expression of Phytophthora resistance. (iii) They have applied a phosphoproteomics approach in order to identify the soybean proteins that are differentially phosphorylated following P. sojae infection. (iv) Recently they have applied the 454-sequencing technology in determining steady state transcript levels of both soybean and P. sojae genes in the resistant and susceptible responses of soybean following P. sojae infection. Bhattacharyya expects that through application of the multiple molecular biological approaches in studying this model plant-pathogen interaction they will be able to develop novel biotechnological strategies to fight not only P. sojae, but also other soybean pathogens. He hopes that their effort will significantly contribute towards securing sustainable productivity of this very important legume crop. |
|
Bhattacharyya Lab is also engaged in studying the sudden death syndrome (SDS) in soybean. SDS is an emerging disease caused by the fungal pathogen, Fusarium virguliforme. The Bhattacharyya group has recently purified a phytotoxin produced by the pathogen. This toxin causes symptoms similar to foliar SDS. They have developed monoclonal antibodies against the purified toxin. Recently they have cloned the toxin gene. Bhattacharyya Lab is intertested in understanding the mechanism used by the pathogen to cause SDS. Earlier they have shown that only in presence of light the toxin can cause foliar SDS and rapid degradation of Rubisco large subunit. They utilized the toxin protein in isolating the host factor(s) that is involved in carbondioxide transport. They have shown that expression of a synthetic single chain variable fragment anti-toxin antibody gene resulted in enhanced foliar SDS resistance in transgenic soybean plants. 
Bhattacharyya group is also involved in understanding the nonhost resistance mechanism of Arabidopsis against the soybean pathogen, P. sojae. They have developed over 3,000 individual M2 families by treating pen1 mutant with ethyl methane sulfonate. The screened the mutant population by infecting with P. sojae spores. They have been able to identify several putants some of which are already fixed to homozygosity. Two of the Pss genes, Pss1 and Pss30, were mapped. They have shown Pss1 confers immunity not only against P. sojae, but also against the fungal pathogen F. virguliforme that causes SDS in soybean. 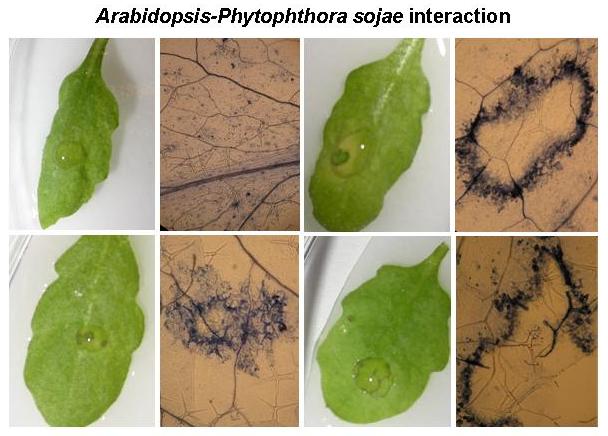
Bhattacharyya group has also been engaged in cloning an active transposable element from soybean. They have collaborated with Dr. Reid Palmer in cloning this element. Previously, Dr. Palmer has generated several lesion mimic root mutants by using this element. Bhattacharyya group is interested in cloning the lesion mimic mutant gene using this transposable element. The Bhattacharyya lab, in collaboration with Palmer and Sandhu Laboratories, has isolated a soybean gene involved in male and female gamete developmen by using this elementt. 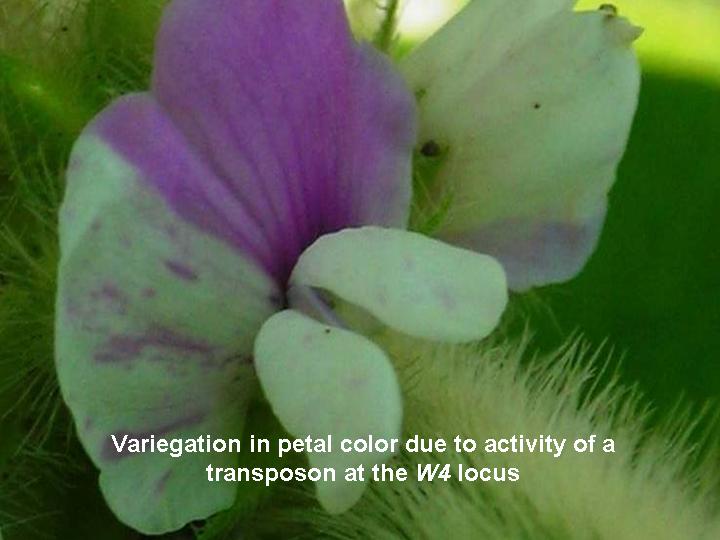 |