The overall goal of the proposed project is to increase the efficiency of crop breeding programs by developing and deploying improved genomic selection strategies that rely on improvements in the selection and mating steps.As a consequence of growing populations, changing diets, and the challenges of climate change, agricultural systems must produce more with less. More means greater demand for agricultural products such as food, feed, energy and fiber. Less means reduced agricultural inputs (at least on a per output basis) such as water, fertilizer, pesticides and a reduced environmental footprint, all on less land. A key tool for making agriculture systems more productive, sustainable and resilient is genetic improvement via breeding.Selection based on favorable (phenotypic selection) traits was central to the domestication of crops and has been used successfully in plant breeding for thousands of years. Recurrent selection is a special case of phenotypic selection that refers to a type of population improvement which includes the following key steps: phenotyping (evaluation), selection, and mating.Phenotyping involves measuring trait values of a population, such as grain and biomass yield, flowering time and color, etc. This step is often time-consuming/labor intensive and many phenotypes cannot be accurately scored without replicated tests.Selection involves the identification of those individuals within the population that exhibit the most favorable trait values.Mating occurs following selection and selected lines are typically randomly or arbitrarily mated to create a new population for phenotyping and selection. Little research has been conducted on the effects of other mating strategies.The phenotyping step can be costly, time consuming, logistically challenging, and/or destructive because it often involves a large number of individuals and many factors that can affect the measurements. Genomic selection (GS), which relies on efficient, high-throughput genotyping technologies (to determine the genetic composition of plants), enables breeders to predict the phenotype of plants based on their genotypes using advanced statistical models and a population of plants of known phenotypes used to “train” the prediction model. This new approach has transformed breeding, because it eliminates or reduces the need for phenotyping.Even moderately accurate genomic prediction can result in substantial cost saving and improve the rate of genetic gain in breeding programs. Previous literature has focused on three areas: the first is to improve the accuracy of phenotype prediction, upon which selection is based; the second is to define other quantitative indicators in lieu of the predicted phenotype to reflect the fitness of the lines from the genetics perspective and their likelihood to produce superior progeny; the third is the selection of an optimal training population that maximizes the accuracy of the prediction model when only a limited number of plants can be phenotyped. Although genomic prediction has transformed breeding, the selection and mating steps have not received equal attention and typically remain the same as in traditional phenotypic selection. Optimizing these two steps is a focus of the proposed project.To design optimal selection and mating strategies and to understand their interactions with other aspect of breeding programs we will make use of simulations within a framework of operations research, which deals with the application of advanced analytical methods to help make better decisions. Significantly, we will also develop user-friendly tools to allow breeders to optimize these parameters for their own breeding scenarios. The new selection and mating strategies that we propose will be designed using advanced mathematical programming and optimization techniques, fully tailorable to reflect user preferences with respect to the trade-offs among cost, time, and probability of success.
Low-cost nitrate sensors to populate genotype-informed yield prediction models for next generation breeders
Our civilization depends on continuously increasing levels of agricultural productivity, which itself depends on (among other things) the interplay of crop varieties and the environments in which these varieties are grown. Hence, to increase agricultural productivity and yield stability, it is necessary to develop improved crop varieties that deliver ever more yield, even under the variable weather conditions induced by global climate change, all the while minimizing the use of inputs such as fertilizers that are limiting, expensive or have undesirable ecological impacts. By coupling a network of innovative, low-cost nitrate sensors across multiple environments within the heart of the corn belt and advanced cropping systems modeling (APSIM, the most widely used modeling platform), the proposed research will enhance our understanding of and ability to predict yield and Genotype x Environment interactions. The integration of nitrate (N) dynamics into this model is expected to greatly increase the accuracy of its predictions. Because we will also integrate genotypes into this model, the proposed research outlines a new and innovative approach for breeding crops that exhibit increased yields and yield stability. It will be possible to readily translate this approach to other crops. By generating data on nitrate concentrations in soil and in planta at unprecedented spatial and temporal resolution at multiple sites with different soil characteristics and weather, the proposed research will also improve our understanding of N cycles in both the soil and plant. Although essential to plant growth and high yields, when over-applied N can result in a variety of serious negative externalities, some of which are currently the subject of high-impact litigation in Iowa. Project outcomes have the potential to provide guidance to farmers about how to apply sufficient but not excessive amounts of N fertilizer, resulting in both economic benefits to farmers and positive environmental externalities.Our focus on creating a new approach to breeding for yield stability meets the USDA sustainability goals to “satisfy human food and fiber needs” and “sustain the economic viability of farm operations”. Our focus on nitrogen meets the USDA sustainability goals to “enhance environmental quality” and to “make the most efficient use of nonrenewable resources…and integrate, where appropriate, natural biological cycles and controls”. More specifically, this proposal addresses the NIFA-Commodity Board co-funded priority for “development and application of tools to predict phenotype from genotype” and the “the development of high-throughput phenotyping equipment and methods”.
Genetic networks regulating structure and function of the maize shoot apical meristem
The shoot apical meristem (SAM) is responsible for development of all above ground organs in the plant. SAM structure and function correlates with agronomically-important adult traits in the maize plant, and is also affected by planting density and shade stresses induced by agricultural environments. The ultimate goal of this project is to increase understanding of the regulatory networks controlling SAM structure and function and the responses of these networks to environmental stresses. The specific objectives are to: 1) describe the SAM allometric space in maize and its relatives using nanoscale computer tomographic scanning to provide 3-dimensional images of the phenotypic diversity of SAM structure and identify adult plant traits correlated with SAM structure; 2) identify differentially expressed genes in SAM size/shape outliers and mutants with abnormal SAM structures and generate a co-expression network of key genes implicated during SAM structure and function; 3) perform quantitative genetic analyses to identify specific variations within genes that correlate with variations in SAM structure/function and adult plant traits, and test functions of 40 key genes using reverse genetic aaproaches; 4) analyze the shade avoidance response and its effects on SAM structure and function; and 5) investigate epigenetic changes of SAM functional domains in response to shade avoidance using novel protocols that distinguish the stem cell organizing regions from the organogenic domains in the maize SAM.
These studies will provide the framework for scientific training and the public release of original data. Undergraduates at Truman State University, a small liberal arts institution, will be trained in morphological and LM-RNAseq analyses of maize mutants. REU students and undergraduates enrolled in Plant Physiology courses at Cornell University will participate in physiological experiments. This project will generate extensive transcriptomic data and vector constructs for tissue-specific epigenetic analyses which will be available to the scientific research community. Molecular markers and phenotypic data for diverse maize lines will be supplied to Panzea. Genetic mapping associations, physiological shade-avoidance response data, transcriptomic and phenotypic data will be curated at MaizeGDB, and seed stocks for maize shoot mutants and SAM size variants will be released through the Maize Genetics Cooperation Stock Center.
Hierarchical Modeling and Parallelized Bayesian Inference for the Analysis of RNAseq Data
This proposal focuses on the development of hierarchical models and parallelized Bayesian inference for the analysis of RNA sequencing (RNAseq) data. Special emphasis is placed on gene expression profiling of parental inbred lines and their hybrid offspring for the discovery of key genes underlying heterosis, the genetic phenomenon otherwise known as hybrid vigor. The project will be led by a collaborative team of researchers with expertise in the analysis of high-dimensional gene expression data, Bayesian inference, bioinformatics, biology, computational methods, genetics, genomics, and statistics. The proposed research provides new tools for the analysis of high-dimension and low-sample-size count data generated by RNAseq technology. Hierarchical modeling allows for flexible information sharing across dimensions to extract as much information as possible from data. Parallel methods for Bayesian inference harness the power of modern computing to produce comprehensive results in a timely manner. Specific methods will be developed for (i) the identification of genes that exhibit expression heterosis, (ii) the detection of expressed and non-expressed genes, and (iii) the discovery of differential allele usage in hybrids. These methods will provide a deeper understanding of the molecular mechanisms of heterosis and lead to the discovery of key genes whose expression patterns provide hybrids with advantages over their parents. This information can be used to efficiently predict which of thousands of possible crosses will result in top performing hybrids. In addition to the specific methods mentioned above, hierarchical generalized linear models for the simultaneous analysis of tens of thousands of response variables will be developed. This work will permit the analysis of RNAseq data from complex designs with multiple sources of variability and will greatly extend the range of applicability for the funded research to encompass a variety of challenges in high-dimensional data analysis.
Public Health Relevance: The proposed work will provide medical researchers with advanced tools for studying the functions of genes in complex biological systems. The enhanced understanding of gene functions obtained with the developed tools can deepen understanding of diseases and lead to new treatments for the improvement of public health.
NRT-DESE: P3 – Predictive Phenomics of Plants
NRT- DESE: Predictive Phenomics of Plants (P3)
New methods to increase crop productivity are required to meet anticipated demands for food, feed, fiber, and fuel. Using modern sensors and data analysis techniques, it is now feasible to develop methods to predict plant growth and productivity based on information about their genome and environment. However, doing so requires expertise in plant sciences as well as computational sciences and engineering. This National Science Foundation Research Traineeship (NRT) award to Iowa State University will bring together students with diverse backgrounds, including plant sciences, statistics, and engineering, and provide them with data-enabled science and engineering training. The collaborative spirit required for students to thrive in this unique intellectual environment will be strengthened through the establishment of a community of practice to support collective learning. This traineeship anticipates preparing forty-eight (48) master’s and doctoral students, including twenty-eight (28) funded doctoral students, with the understanding and tools to design and construct crops with desired traits that can thrive in a changing environment.
Understanding how particular genetic traits result in given plant characteristics under specific environmental conditions is a core goal of modern biology that will facilitate the efficient development of crops with commercially useful characteristics. Plant characteristics are influenced by genetics and a wide range of environmental factors, including, for example, rainfall, temperature and soil types. Developing methods to effectively integrate these diverse inputs that take advantage of existing biological, statistical, and engineering knowledge will be a key area in this research and training program that will bring together faculty from eight departments. Trainees will engage in cutting-edge research and development areas involving direct data collection and analysis from living plants, including sensor development, high throughput robotic technology, and biological feature extraction through image analysis. This traineeship will use the T-training model to provide students with training across a broad range of disciplines while developing a deep technical expertise in one area. This expertise, in combination with soft skills development, will enable the trainees to work across organizational and cultural boundaries as well as scientific disciplines. To develop understanding of how to share knowledge with diverse groups, the program will provide students with training beyond traditional coursework and research through activities that will develop advanced communication and entrepreneurship skills. Additionally, internship opportunities in industry, national labs, and other settings will equip trainees to choose among the diverse career paths available to scientists and engineers.
The NSF Research Traineeship (NRT) Program is designed to encourage the development and implementation of bold, new, potentially transformative, and scalable models for STEM graduate education training. The Traineeship Track is dedicated to effective training of STEM graduate students in high priority interdisciplinary research areas, through the comprehensive traineeship model that is innovative, evidence-based, and aligned with changing workforce and research needs.
Parallel Algorithms and Software for High-Throughput Sequence Assembly
High-throughput next-generation DNA sequencing technologies (NGS) are causing a major revolution in life sciences research by allowing rapid and cost-effective sampling of genomes and transcriptomes (expressed genomic sequences). Assembly of genomes and transcriptomes from billions of such randomly sampled sequences is an important problem in computational biology. While significant strides have been made, much work remains in addressing the diverse and rapidly emerging platforms, improving assembly quality, and scaling to both large-scale data sizes and large genomes.
This project will harness the power of high performance computing to develop effective solutions for sequence assembly. It will lead to the development of scalable, efficient parallel algorithms and a parallel integrated software framework for genome and transcriptome assembly. The project seeks to advance the state of the art by targeting important unsolved problems such as hybrid assembly of sequences from multiple NGS platforms, making fundamental algorithmic advances to improve assembly quality, and conducting an in-depth effort at parallel algorithms development for the entire gamut of problems that arise in connection with assembly. It will be carried out by an interdisciplinary team of investigators, in partnership with leading NGS manufacturers and academicians involved in large plant genome sequencing projects.
The project will lead to the release of a scalable parallel software package for sequence assembly that will be made available to the scientific community. Postdoctoral and graduate students will be trained in computer science driven interdisciplinary research and in writing efficient high performance computing software. The project will influence curriculum development and will lead to educational materials in bioinformatics for next-generation sequencing.
“Transgenic Approaches In Managing Sudden Death Syndrome In Soybean”
Our long-term goal is to create soybean cultivars resistant to soybean sudden death syndrome (SDS). Soybean is one of the world’s most valuable crops and the U.S. is the world leader in soybean production. In 2010 the U.S. soybean crop value was over $38.9 billion. Soybean suffers yield suppression from various biotic stresses, including SDS, which in 2010 caused losses valued at $0.82 billion.
The transdisciplinary project team consists of experts from states and countries where soybean is an important crop: Iowa, Illinois, Indiana, Brazil and Argentina.
Objectives are:
- Apply transgenic approaches to i) suppress fungal growth through RNA interference, ii) generate antibodies against toxins that induce SDS, iii) overexpress maize carbonic anhydrase and Arabidopsis nonhost resistance genes to enhance SDS resistance, and iv) express an effector protein under regulation of a F. virguliforme-infection inducible promoter.
- Incorporate successful transgenes and novel SDS resistance genes into elite soybean lines.
- Determine the use of transgenes against a diverse collection of SDS pathogen isolates.
- Evaluate economic and social impact of transgenes.
- Provide education and research experience to 6-12 grade teachers and undergraduate minority students.
- Educate stakeholders and youth via extension programming on the use of transgenic technology to ensure sustainable soybean yield.
Outcomes include enhanced profitability for soybean growers and a secure, sustainable supply of soybean for the 21st century.
Sorghum breeding program for biofuel production
Biofuels are a major contributor to the energy security of the United States, to the economic growth of Iowa and to the reduction of greenhouse gasses emission. The Energy Independence and Security Act (2007) established that 36 billion gallons of biofuels per year had to be produced by 2022. In 2018, 16 billion gallons of ethanol were produced from maize, but maize-based ethanol cannot supply the total demand and it has detrimental implications for food and feed supplies. Therefore, other sources, such as lignocellulosic feedstock, need to be developed.
In 2008, Dr. Salas Fernandez initiated the sorghum breeding program for biofuel production at Iowa State University. The main goal is to conduct research for the development of sorghum germplasm for biofuel production adapted to Iowa. The breeding program is centrally located in Ames, IA, with winter nursery activities in Puerto Rico and three testing locations in Iowa were experimental hybrids are evaluated every year.
Sorghum ethanol yields vary depending on the type of sorghum cultivated. Sweet sorghums can produce 900 gallons/acre, if we consider a standard composition, yields of 16 Tn of dry matter/ha and 90% conversion efficiency. Our yield trials demonstrate that biomass sorghum can produce up to 1,120 gallons/acre as a lignocellulosic feedstock, considering our highest yields of 35 Tn dry matter/ha, a standard composition and a 90% conversion efficiency. Therefore, sorghum could become the preferred bioenergy crop, considering its high yield potential and the additional benefit of low input use, since it requires less nitrogen and water than corn.
In addition to the ethanol production from corn grain, commercial lignocellulosic biorefineries are processing corn-derived biomass, a dry and low volume feedstock. Recent evidence suggests that the greenhouse gas (GHG) benefit of cellulosic ethanol from corn stover is marginal relative to fossil fuel production. Therefore, sorghum biomass could be used as a new source of cellulosic fuel with net GHG savings relative to corn stover. The advantage will be to produce biofuel with a reduced environmental impact and without competing for food production. Considering that storage and transportation of high-tonnage biomass with high water content is a major limitations to the development of a strong bioeconomy, anaerobic digestion (on-farm or at centralized facilities) is considered a promising conversion technology to generate biogas. The ISU sorghum breeding program is developing novel germplasm for these goals and processing methods.
Photosynthesis in sorghum under non-stress, cold and drought stress
Carbon assimilation through photosynthesis is the basis of crop productivity. However, increases in crop yield achieved in the last 50 years have not been attributed to changes in photosynthetic capacity. The complex genetic architecture of C assimilation and the lack of correlation between grain yield and photosynthesis were the most important arguments to postpone significant investments in this scientific area. The advancement of “omics” technology, high-throughput phenotyping methods and biofuels has significantly changed the paradigm. Considering there is a direct association between photosynthetic efficiency and biomass yield, the discovery and exploitation of the genetic architecture controlling C assimilation could have a significant impact on biomass yield for biofuel production. Dr. Salas Fernandez and her team are investigating genes/alleles associated with higher leaf photosynthetic capacity under non-stress, cold and drought stress using both field and controlled condition experiments.
Several genomic regions associated with gas exchange and chlorophyll fluorescence parameters were discovered and are currently being validated. These studies have demonstrated the existence of natural genetic variation in C fixation that could be exploited to breed for superior germplasm.
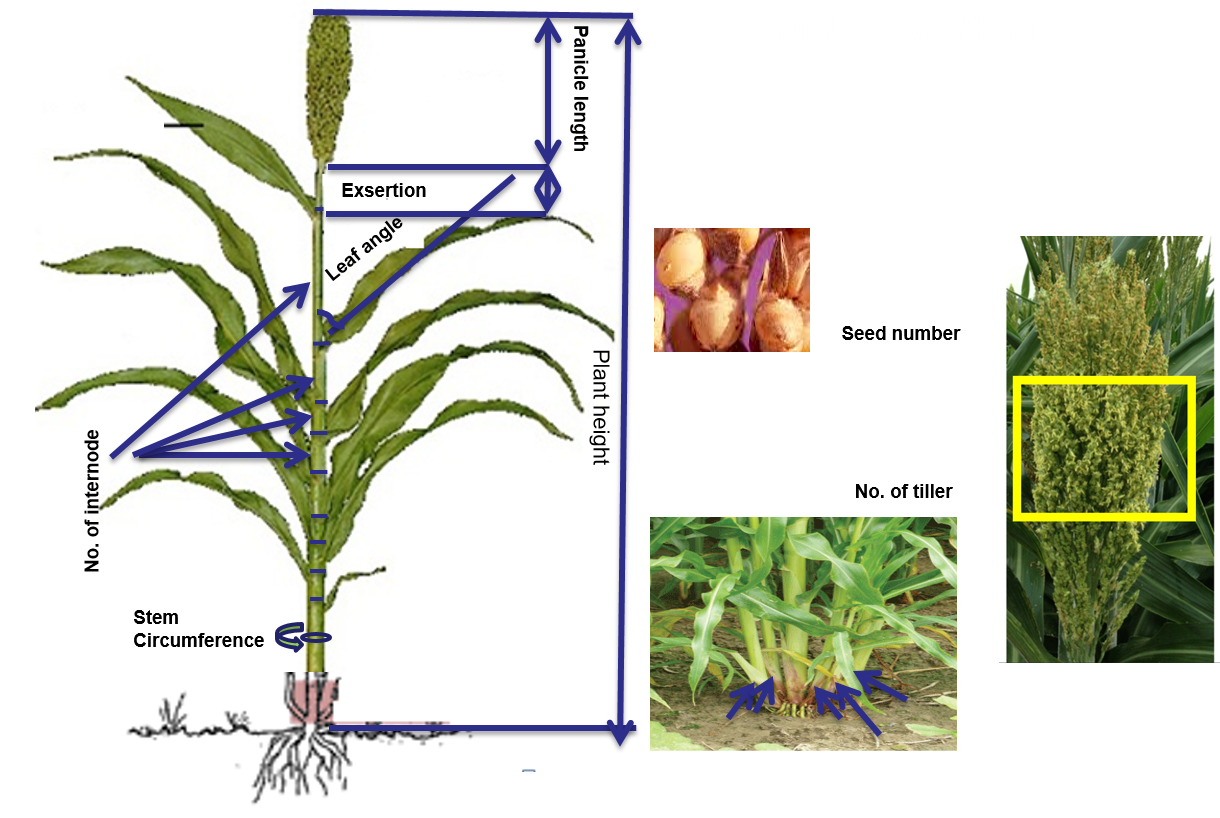
Plant architecture
Several hormones are involved in the biochemical and physiological responses that determine plant architecture characteristics highly correlated with biomass yield such as plant height, leaf angle, stem diameter, tillering, number of florets, etc. Brassinosteroids, gibberellins and auxins have the strongest impact without severe undesirable pleiotropic effects. Identifying genes involved in biosynthetic and signaling pathways of these groups of hormones and the effects of alternative alleles will reveal the allelic combination to obtain a particular plant type. Sorghum germplasm offers a vast genetic diversity to dissect plant architecture traits and identify genes/alleles controlling specific characteristics.
We have conducted genome-wide and candidate gene association studies to characterize the genetic architecture of plant height, stem diameter, leaf angle, exsertion, panicle lenght, internode number, flowering time, seed number per panicle and tiller number using the Sorghum Association Panel. We have discovered genomic regions and candidate genes that are currently being validated. With the need to produce more food, feed and fuel in the same or smaller area, and considering climate change, manipulating genes to create desirable plant types in a shorter period of time will be essential in breeding programs. Sustainable production of biofuel will also require using fewer inputs, in more marginal lands, and therefore producing a specific sorghum plant for that production system will be very valuable as well.
RESEARCH-PGR: A Genome-level Approach to Balancing the Vitamin Content of Maize Grain
This collaborative research project is directed at identifying a subset of the ~40,000 genes in the corn genome that work together to determine the levels of five essential and limiting dietary vitamins in kernels: vitamin E and the four B vitamins, B1 (thiamin), B2 (riboflavin), B3 (niacin) and B6 (pyridoxine). By combining approaches similar to those used in the Human Genome project, the researchers will identify alleles, special variations in these “vitamin” genes, and learn how to put them together to generate high amounts of vitamins in corn kernels. An important outcome of this research will be the knowledge by which to enhance these micronutrient levels in corn kernels such that diets in which maize is a major component provide a balanced nutritional content. Such direct translation of these findings will be the eventual incorporation and fixation of identified alleles in maize breeding programs that are favorable for the increased levels of vitamins E and B to enhance the food and feed supply chain. In addition, this research will provide guiding principles for parallel efforts in other agricultural crops and thus enable predictive breeding and metabolic engineering of more nutritious crops worldwide. Finally, integration of research with education within the project will permit training of the next generation of plant scientists with knowledge of plant genetics, breeding, genomics, biochemistry, and bioinformatics.
This project seeks to leverage the tremendous genetic and genomic tool sets developed in maize the past decade to advance and accelerate our fundamental understanding of the genes, alleles and genetic mechanisms controlling synthesis and accumulation of vitamins that are limiting in maize grain and hence result in vitamin deficiencies in maize-based diets: four B vitamins (B1, thiamine; B2, riboflavin; B3, niacin; B6, pyridoxine) and vitamin E. This project brings together a team of scientists with divergent but complementary knowledge and skills that together will allow the genes, alleles and underlying mechanisms controlling these nutritional traits to be elucidated and the knowledge deployed on a global scale. Specific objectives are to (i) perform genome-wide association studies with the maize Ames inbred line panel (n~2,000) to identify and resolve quantitative trait loci (QTL) controlling accumulation of these micronutrients; (ii) assess the role of rare alleles by constructing and analyzing segregating F2 populations derived from Ames lines that are extreme outliers for traits; (iii) determine the contribution of expression QTL and presence-absence variants (PAVs) to vitamin composition using whole transcriptome sequencing data obtained from grain 24 days after pollination in 500 inbred lines that represents the phenotypic variation of the Ames panel; and, (iv) perform genomic prediction with the Ames panel to accelerate the efficiency of breeding improved grain micronutrient composition in developing countries. The broader impacts of this project to the broader scientific community and public will be ensured through a set of coordinated activities that engage students, postdoctoral associates, scientists and the public. Data and biological resources generated in this project will be made accessible to the community. Data will be disseminated through publications, project websites and long-term repositories such as the NCBI’s SRA and MaizeGDB.
FACT: International workshop on Machine Learning for Cyber-Agricultural Systems
PI – Soumik Sarkar; Co-PI – Arti Singh, Baskar Ganapathysubramanian, Asheesh Singh
The proposers in collaboration with Prof. Masayuki Hirafuji & team at the University of Tokyo, are organizing the First International workshop on Machine Learning for Cyber-Agricultural Systems (MLCAS 2018) on October 24, 2018 as a part of the Asia-Pacific Federation for Information Technology in Agriculture and the World Congress on Computers in Agriculture (AFITA/WCCA) being held from October 24, 2018 to October 26, 2018 that will be collocated at the Victor Menezes Convention Centre at the Indian Institute of Technology, Bombay (IITB) in Mumbai, India